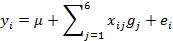Abstract
Female fertility is an economically important trait in the dairy industry, and the fertility selection index has been developed as a method of including female fertility in the breeding goals of this industry. This index considers a combination of factors, including days open, number of inseminations per lactation, success after first insemination, and pregnancy within 70 d, 90 d, and 110 d after parity. Based on a genome-wide association study of the fertility selection index using 442 Holsteins, we found that the index is influenced by a variation in the thioredoxin fold region of the family with the sequence similarity 213, member A (FAM213A) protein. FAM213A is a CXXC motif-containing peroxiredoxin 2-like protein that regulates cellular redox status. A replacement of isoleucine with valine in FAM213A was associated with poor fertility in cows. The overexpression of FAM213AVal in bovine endometrial epithelial cells reduced reactive oxygen species to a lesser extent relative to the overexpression of FAM213AIle and caused a decrease in cyclooxygenase-2 expression. Downregulation of cyclooxygenase-2 led to a decline in prostaglandin E2, which is critical for implantation because it protects the conceptus from the maternal immune system. Cows with FAM213AVal showed lower levels of prostaglandin E2 than did cows with FAM213AIle, suggesting that cows with FAM213AVal are less fertile than cows with FAM213AIle because of their reduced uterine environment. Thus, the present study found that FAM213A unexpectedly modulates female fertility in cattle.
Author Contributions
Academic Editor: Sekhar Kambakam, Iowa State University
Checked for plagiarism: Yes
Review by: Single-blind
Copyright © 2016 Mayumi Sugimoto, et al
 This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Competing interests
The authors have declared that no competing interests exist.
Citation:
Introduction
Dairy production depends on the frequency at which cows conceive; thus, female fertility is essential for this industry. To improve overall female fertility, the dairy industries in certain countries have combined several reproductive components into one overall index: the fertility selection index (SI) 1, 2, 3, 4. In Japan, the SI consists of the estimated breeding values (EBVs) for days open (DO), the number of inseminations per lactation (NI), success after first insemination (SFI), and pregnancy within 70 d (P70), 90 d (P90), and 110 d (P110) after parity 5. The first two traits are continuous traits, whereas the other four traits are binary traits that show either success or failure after insemination. The genetic correlations among these reproductive traits are not necessarily ± 1.0 1, 3. Therefore, a principal components analysis is used to estimate weights for each trait and combine them into the SI to allow for the selection of these six fertility traits simultaneously.
Several groups have already conducted genome-wide association studies (GWASs) for SI using Danish Jersey 4, Nordic Red 6, and Italian Holstein 7. These studies revealed some single nucleotide polymorphisms (SNPs) associated with both SI and individual traits included SI, suggesting that important fertility traits for selection in the population would generate the signal. Thus, studying the genetics of the SI using a GWAS may be helpful for understanding the underlying biological mechanisms and improving the general fertility level of the population.
Materials and Methods
Samples
We collected blood from 7,034 Holstein cows and evaluated their SI 5. The mean SI was 0, and cows with a SI lower than -1.35 were in the 5th percentile, whereas cows with a SI higher than 1.46 were in the 95th percentile of the study population (Figure 1A). We selected samples from 212 cows with an SI lower than -1.35 as well as samples from 230 cows with an SI higher than 1.46.
Figure 1.Fertility selection index (SI) is associated with SNP BovineHD2800009796. A. The distribution of SI values among 7,034 cows. B. A quantile-quantile plot of the GWAS for the SI. The equiangular line (black line) is included in the plot for reference purposes. The dashed horizontal line indicates the threshold for genome-wide significance (assuming a Bonferroni correction) for the 782,696 single nucleotide polymorphisms (SNPs) tested. BovineHD2800009796 is the SNP most significantly associated with the SI on a genome-wide level.
Whole-Genome Scan
We genotyped 176 low SI samples and 174 high SI samples using the BovineHD BeadChip (Illumina, San Diego, CA, USA) for a total of 782,696 SNPs. SNPs were excluded if they deviated significantly from Hardy-Weinberg equilibrium (p < 0.001), if they had low minor allele frequency (< 0.01), or if they had a call rate < 95 %. We have previously genotyped 36 low and 56 high SI samples using the BovineSNP50 BeadChip (Illumina) 8 and combined these genotypes into the BovineHD data by treating these 92 samples missed genotype for HD-specific SNPs. We conducted an association analysis using EMMAX software 9.
Identification of Novel SNPs
Based on the Oct. 2011 Bos taurus draft assembly 10 (Baylor Btau_4.6.1), each of the exons, 2 kb of the 5’ untranslated regions (UTRs) and 2 kb of the 3’ UTRs of the genes located in the associated regions were amplified by polymerase chain reaction (PCR) and sequenced. We defined the genome-wide regions that included significant SNPs as well as their neighboring SNPs with r2valuesgreater than 0.2 as the associated region because a linkage disequilibrium decayed rapidly until r2 reached 0.2 in cattle 11. The r2values were calculated by a linkage disequilibrium analysis using PLINK software 12. The primers for each gene and the samples used to compare the sequences are shown in Tables A and B in the Supplemental data, respectively.
Allelic Substitution Effects
We genotyped 2,682 cows and 4,165 bulls for the family with sequence similarity 213, member A (FAM213A), plakophilin 2 (PKP2), cortactin-binding protein 2 N-terminal like (CTTNBP2NL), SET domain containing 6 (SETD6), calcium channel, voltage-dependent, beta 2 subunit (CACNB2), andunc-5 homolog C (UNC5C). FAM213Awas identified as a gene that was associated with the SI in the present study, whereas PKP2, CTTNBP2NL, SETD6, CACNB2 and UNC5C have previously been identified as genes associated with the conception rate (CR) 8, 12. The EBVs of cows and bulls with these six traits included in the SI were estimated by a multiple-trait animal model using 1,881,898 records. The EBVs of cows and bulls in relation to the CR were estimated by a threshold repeatability animal model using 3,428,666 values after first parturition. The data were collected between January 1990 and September 2015 by the dairy herd improvement program of Hokkaido, Japan. The allelic substitution effects of these genes were determined using the following equation:
where  = the deregressed estimated breeding value (dEBV) 13 of animal i (= 1, 2, …, n) for the SI, DO, NI, SFI, P70, P90, P110, or CR;
= the deregressed estimated breeding value (dEBV) 13 of animal i (= 1, 2, …, n) for the SI, DO, NI, SFI, P70, P90, P110, or CR; 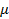 = the general average value of the population;
= the general average value of the population; 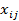 = the genotype covariate (coded as 0 or 2 for the two homozygotes and 1 for heterozygotes) of gene j in animal i;
= the genotype covariate (coded as 0 or 2 for the two homozygotes and 1 for heterozygotes) of gene j in animal i; 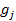 = the random regression coefficient representing the allelic substitution effect for gene j; and
= the random regression coefficient representing the allelic substitution effect for gene j; and 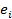 = the random residual effect for the value of animali. We performed the analyses with the SAS MIXED procedure.
= the random residual effect for the value of animali. We performed the analyses with the SAS MIXED procedure.
Real-Time PCR
RNA was extracted from individual samples of bovine brain, kidney, liver, lung, mammary gland, muscle, ovaries, pancreas, stomach, and uterine tissue or from transfected cells using TRIzol reagent (Life Technologies, Carlsbad, CA, USA). Real-time PCR was conducted with an ABI 7900HT Sequence Detection System using the comparative Ct method and glyceraldehyde-3-phosphate dehydrogenase (GAPD) as an internal control (Life Technologies). The primers used in these assays are shown in Table C in the Supplemental data.
Generation of Overexpression Constructs
We generated bovine FAM213A by PCR with the gene’s respective forward and reverse primers (Table D in Supplemental data. These PCR products were further amplified via PCR with the forward-2 and reverse-2 primers (Table D in the Supplemental data) for cloning into a pCAGGS (R-N) vector 14 using an In-Fusion Advantage PCR Cloning Kit (Takara Bio Inc., Shiga, Japan).
Cell-Viability Assay
To check the redox activity in cells transfected with bovine FAM213A, we performed cell-viability assays. Bovine endometrial epithelial cells (BEnEpCs; Cell Applications, San Diego, CA, USA) were plated and transfected. H2O2 was diluted in PBS and further diluted in culture medium to the indicated concentrations. Cells were incubated in medium containing H2O2 for 24 h, and cell viability was evaluated with the MTT-based Cell Growth Determination Kit (Sigma-Aldrich, Saint Louis, MO, USA) according to the manufacturer’s instructions. The amount of MTT formazan produced was determined by measuring absorbance with a microplate reader (Bio-Rad, Hercules, CA, USA) at a test wavelength of 570 nm and a reference wavelength of 655 nm. The p-value was calculated using Student’s t-test.
Measurement of the GSH/GSSG Ratio
We also measured the GSH/GSSG ratio to compare the cellular redox status in cells transfected with bovine FAM213A using the glutathione detection kit (ENZO Life Sciences, Farmingdale, NY, USA) according to the manufacturer’s instructions. In brief, total glutathione (GSHt) and GSSG concentrations were derived from GSH and GSSG standard curves and converted into nanomoles per milligram of protein. The concentrations of reduced GSH were found by subtracting GSSG from GSHt. Finally, the GSH/GSSG ratio was calculated by dividing the difference between GSHt and GSSG concentrations by the GSSG concentration ratio = GSHt – 2 (GSSG)/GSSG. The p-value was calculated using Student’s t-test.
Immunoblotting Analysis
To investigate cyclooxygenase-2 (COX-2) expression in cells transfected with bovine FAM213A, we analyzed immunoblotting. Protein was extracted from transfected cells using NucleoSpin RNA/Protein (Machrey-Nagel, Düren, Germany). The extracted proteins were separated, transferred to a membrane, and blocked. The blots were incubated with an anti-COX-2 (ab15191, Abcam, Cambridge, UK) or anti-actin (A2066, Sigma-Aldrich) antibody and detected with ECL Prime (GE Healthcare, Buckinghamshire, UK). To estimate the ratio of COX-2 to actin, the obtained images were quantified using ImageQuant LAS 4000 (GE Healthcare).
Enzyme Immunoassays
To assess prostaglandin E2 (PGE2) secreted by cells transfected with bovine FAM213A, we conducted immunoblotting analysis. Pyrrolidine dithiocarbamate (PDTC) and N-(2-cyclohexyloxy-4-nitrophenyl)-methane sulfonamide (NS-398) were purchased from Sigma-Aldrich. Before transfection, BEnEpCs were pretreated with 10 µM PDTC or 10 µM NS-398 for 30 min. The concentration of PGE2 released from the transfected BEnEpCs or the bovine serum were assayed using a PGE2 ELISA kit (ENZO Life Sciences) according to the manufacturer’s instructions. The p-value was calculated using Student’s t-test.
Results
Our GWAS identified BovineHD2800009796 as the most significantly associated SNP on chromosome 28 (Figure 1B), and there were no more SNP that may be impacting SI as shown in Graph A in the Supplemental data. The region associated with this SNP included 13 genes (Figure 2A). To detect possible causative polymorphisms in this region, we sequenced all exons and the 5’ and 3’ UTRs of these 13 genes and found 42 novel SNPs (Figure 2A). The positions, the minor allele frequencies, and the genotype counts in the GWAS sample for the novel 42 SNPs in Table E in the Supplemental data. A reanalysis of the newly sequenced SNPs demonstrated that FAM213A (A31G: I11V) was the most significant (Figure 2A). Moreover, we genotyped FAM213A (A31G: I11V) in 2,682 cows and 4,165 bulls and found that the allele substitution effect of FAM213A on the dEBV for the SI was 0.18 (Figure 2B). Cows with the A/A genotype, which codes for Ile/Ile, exhibited a 0.18 higher SI than those with the A/G genotype, which codes for Ile/Val, whereas cows with the G/G genotype, which codes for Val/Val, exhibited a 0.18 lower SI than those with the A/G genotype. FAM213A also had favorable effects on the dEBV for the traits that compose the SI (DO, NI, SFI, P90, and P110). The effects of FAM213A were similar to those of PKP2 (chromosome 5: 82,452,366 bp), CTTNBP2NL (chromosome 3: 33,171,753 bp), SETD6 (chromosome 18: 26,204,767 bp), CACNB2 (chromosome 13: 32,851,153 bp), and UNC5C (chromosome 6: 31,142,900 bp), which have previously been identified to be associated with the CR in the Japanese dairy cow population 8, 12 (Figure 2B). Therefore, FAM213A (A31G: I11V) was the most promising causative SNP on chromosome 28 and had a similar impact on the SI as the five genes previously identified to be associated with CR.
Figure 2.FAM213A SNP is associated with the SI. A. Association signals on chromosome 28 with the SI using plots of the P values from the EMMAX analysis. The black line represents the threshold for genome-wide significance after applying the Bonferroni correction for multiple comparisons. BovineHD2800009796 was the first significantly associated SNP that was detected, whereas FAM213A (A31G: I11V) was the only significant novel SNP. Blue circles represent the positions of the 42 newly sequenced SNPs. Blue lines represent 13 genes located in the associated region. The triangle diagram represents a pairwise linkage disequilibrium (LD) in the associated region, which was visualized using Haploview 35. Red shades indicate strong LD. B. The allelic substitution effects of FAM213A, PKP2, CTTNBP2NL, SETD6, CACNB2, and UNC5C on the deregressed estimated breeding value (dEBV) 14 for the SI traits days open (DO), number of inseminations per lactation (NI), success after first insemination (SFI), pregnancy within 70 d (P70), 90 d (P90), and 110 d (P110) after parity or for the conception rate (CR). n. s.: non-significant. * and **: p < 0.05 and p < 0.01, respectively.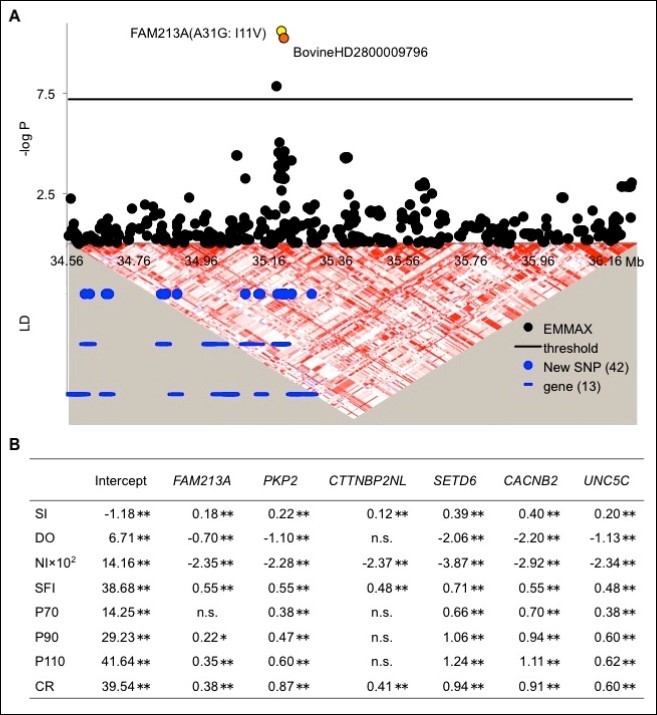
FAM213A protein is a CXXC motif-containing peroxiredoxin (PRX) 2-like protein 16. PRXs and thioredoxins (TRXs) are redox regulatory proteins. PRXs play a protective antioxidant role in cells through their peroxidase activity, whereas TRXs alter the redox state of target proteins by catalyzing the reduction of their disulfide bonds through a CXXC motif. FAM213A has a similar sequence to PRXs and contains a CXXC motif that is shared by many enzymes with redox regulatory functions. Thus our identified two types of FAM213A might modulate reactive oxygen species (ROS) differently. To assess its redox activity, we used BEnEpCs derived from bovine uterine tissue because FAM213A is expressed in several bovine tissues including the uterus (Figure 3A). Cell-viability assays using BEnEpCs transfected with FAM213A revealed that oxidative stress induced by H2O2 treatment decreased cell viability in a dose-dependent manner and indicated that cells with FAM213AVal had increased viability relative to that of FAM213AIle in the presence of 0.2 mM H2O2 (Figure 3B). Consistent with the results of the cell-viability assay, the GSH/GSSG ratio, a proxy for the cellular redox status, was increased in BEnEpCs transfected with FAM213AVal to a greater extent than in BEnEpCs transfected with FAM213AIle (Figure 3C). Furthermore, the GOR prediction for a protein’s secondary structure 17 revealed that FAM213AVal would likely have a shorter b-sheet than FAM213AIle (Figure 3D). These results suggest that the activity of FAM213AVal shifts conditions towards a more reduced environment than does FAM213AIle and indicate that this single amino acid substitution has an impact on the function of this protein.
Figure 3.Overexpression of FAM213AVal reduced reactive oxygen species (ROS) to a lesser extent relative to the overexpression of FAM213AIle in bovine endometrial epithelial cells (BEnEpCs). A. FAM213A expression levels in bovine tissues as determined via real-time PCR. B. Relative cell viability of FAM213AIle or FAM213AVal transfected in BEnEpCs in the presence of H2O2. The data are presented as the mean ± SEM (n = 3). The p-value was calculated using Student’s t-test. *: p < 0.05. C. The relative GSH/GSSG ratio in BEnEpCs transfected with FAM213AIle or FAM213AVal in the presence of 0.2 mM H2O2. D. Schematic illustration of the GOR-predicted protein secondary structure for the partial amino acid sequences of FAM213A. Purple lines represent the b-sheet and green curves represent a-helices.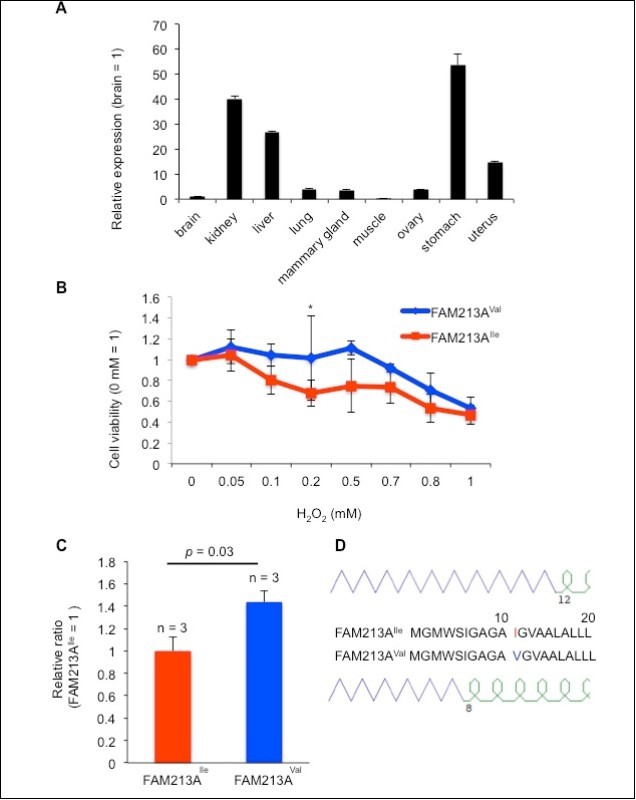
ROS are important factors for fertility. In the uterus, the establishment of a maternal-fetal connection is associated with an increase in ROS 18. Although ruminants have different types of placentation relative to humans or mice, ROS fluxes occur before implantation in all of these mammals 19. Increases of ROS production by peripheral bovine lymphocytes have been reported as pregnancy progresses 20. ROS triggers the NF-κB pathway, which enhances COX-2 expression 21, 22, suggesting that the more reduced environment created by FAM213AVal might decrease COX-2 expression. To explore this possibility, we conducted real-time PCR using RNA extracted from BEnEpCs transfected with FAM213A. As expected, BEnEpCs transfected with FAM213AVal expressed less COX-2 than cells transfected with FAM213AIle (Figure 4A). Moreover, pretreatment of BEnEpCs with a selective inhibitor of NF-κB, 10 µM PDTC, significantly repressed COX-2 expression (Figure 4A). We found the same result in the level of protein expression (Figure 4B). Thus, the more reduced environment created by FAM213AVal appears to decrease COX-2 expression through effects on the NF-κB pathway.
Figure 4.Overexpression of FAM213AVal in BEnEpCs resulted in less cyclooxygenase-2 (COX-2) expression compared with cells overexpressing FAM213AIle. A. The relative COX-2 expression of BEnEpCs transfected with FAM213AIle or FAM213AVal in the absence or presence of pretreatment with 10 µM pyrrolidine dithiocarbamate (PDTC) before treatment with 0.2 mM H2O2. The data are presented as the means of 3 experiments ± SEM. The p-value was calculated using Student’s t-test. B. Representative immunoblots with an anti-COX-2 or anti-actin antibody from BEnEpCs transfected with FAM213AIle or FAM213AVal in the absence or presence of pretreatment with 10 µM PDTC before treatment with 0.2 mM H2O2. Relative ratios of COX-2 to actin (FAM213AIle = 1) are shown.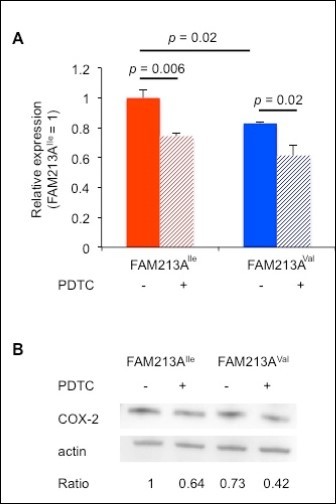
A COX-2 deficiency in mice results in implantation defects 23, and one of the products of COX-2, PGE2, can correct these implantation defects in COX-2-deficient mice 24. Therefore, decreased COX-2 expression might reduce the production of PGE2 to levels that are inadequate for implantation. Our enzyme immunoassays revealed that BEnEpCs transfected with FAM213AVal released less PGE2 compared with cells transfected with FAM213AIle (Figure 5A). Furthermore, pretreatment of BEnEpCs with a selective inhibitor of COX-2, 10 µM NS-398, or a selective inhibitor of NF-κB, 10 µM PDTC, significantly repressed PGE2production (Figure 5A). Additionally, the serum concentrations of PGE2 of cows carrying FAM213AIle/Ile were higher than those of cows carrying FAM213AVal/Ile or FAM213AVal/Val (Figure 5B). Therefore, the reduced uterine environment created by FAM213AVal decreased PGE2production through effects on the NF-κB/COX-2 pathway, which might decrease the SI of cows carrying FAM213AVal.
Figure 5.Cows with FAM213AVal showed lower levels of prostaglandin E2 (PGE2) compared with cows with FAM213AIle. A. The concentration of PGE2 released from BEnEpCs transfected with FAM213AIle or FAM213AVal in the absence or presence of pretreatment with 10 µM N-(2-cyclohexyloxy-4-nitrophenyl)-methane sulfonamide (NS-398) or 10 µM PDTC before treatment with 0.2 mM H2O2. The data are presented as the means of 3 experiments ± SEM. The p-value was calculated using Student’s t-test. B. The serum concentrations of PGE2 of FAM213AIle/Ile or FAM213AVal/- cows at approximately day 9 of the estrus cycle.
Discussion
The present study is the first to demonstrate that FAM213A modulates fertility. Scanning the whole genome of 442 Holsteins and analyzing the results with EMMAX identified a novel mutation associated with the SI. Although we have previously identified several genes associated with the CR in the Japanese Holstein population using a principal component analysis approach 8, 13, FAM213A is a novel gene associated with fertility, and it has only previously been reported to modulate osteoclast differentiation in vitro16. One of the reasons for this GWAS result might be that the SI follows a normal distribution, which makes it possible to compare samples belonging to the 5th percentile with samples belonging to the 95th percentile of the population (Figure 1A). A second reason might be that the EMMAX method could correct for severe stratification and reduce the effects of unusual clusters of cows that are significantly larger because of intensive genetic selection and the extensive use of artificial insemination in the dairy industry 25.
However, in the present GWAS, significant SNPs were not observed near the genes previously identified as associated with CR, although the allelic substitution effects of these genes on the SI were significant among the 2,682 cows and 4,165 bulls (Figure 2B) that were analyzed. The SI consists of reproductive traits that have low heritabilities ranging from 0.05 to 0.14 5. In dairy cattle, only DGAT1 and ABCG2 have been identified and verified by multiple studies to have significant effects on fat and protein concentrations, which have the highest heritabilities of all of the traits that are routinely analyzed 26. The heritabilities of these traits range from 0.20 to 0.61 27. Thus, using multiple GWASs to identify and verify the genes responsible for reproductive traits that have low heritabilities might be more difficult than for milk production traits that have high heritabilities.
We found that cows carrying FAM213AVal have a lower SI than cows carrying FAM213AIle. This single amino acid substitution is located in the thioredoxin fold region and predicted to change the length of the b-sheet (Figure 3D) in this region. The thioredoxin fold region contains a b-a-b-a-b-a-b-b-a arrangement 28, and the length of the first b-sheet of the region might be critical for the redox regulatory function of this protein.
The identified single amino acid substitution decreases ROS, which reduces COX-2 expression through the NF-κB pathway (Figure 4) and reduces PGE2 production (Figure 5). It is worth noting that COX-2 expression and PGE2 production are also induced by interferon-tau 29, the pregnancy recognition signal produced by the trophoblast prior to implantation in ruminants, which has been shown to enhance fetal survival when administered to mice 30, 31. Along with the interferon-tau pathway, enhancing COX-2 expression and PGE2 production via the ROS/NF-κB pathway might be important for implantation.
Although we demonstrated effects using BEnEpCs in the present study, FAM213A might also have impacts on different tissues in cows because the SI consists of several reproductive components 5. In the ovary, oocyte maturation requires ROS 32, and FAM213A was found to be expressed in the ovary (Figure 3A). Ovulation is initiated by a surge in luteinizing hormone, which generates ROS 33. After ovulation, the corpus luteum is produced in the ovary, which also generates ROS 34. Moreover, COX-2-deficient mice have demonstrated multiple failures in female reproductive processes, including fertilization, ovulation, and implantation 23. Thus FAM213A might affect multiple reproductive processes through the ROS/NF-κB/COX-2/PGE2 pathway.
Conclusion
The present study investigated the whole genome in samples from 212 cows with a low SI and samples from 230 cows with a high SI, and a significant association was observed between the SI and FAM213A with a single amino acid polymorphism. Further functional studies revealed that this single amino acid polymorphism in FAM213A reduced ROS, thus leading to decreased COX-2 expression and lower PGE2 production. These results indicate that FAM213A plays a role in female fertility in cows.
Acknowledgments
The authors thank K. Maruyama for performing extensive genotyping and S. Sasaki for providing useful discussions.
Supplemental Data:
Primers used to search for SNPs (Table A). Samples used for developing new SNPs (Table B). Primers used for real-time PCR (Table C). Primers used for generating the overexpression constructs (Table D). A quantile-quantile plot of the GWAS for the fertility selection index without BovineHD2800009796 (Graph A). The minor allele frequencies and the genotype counts in the GWAS sample for 42 novel SNPs in chromosome 28 (Table E). The standard error for the allelic substitution effects of FAM213A, PKP2, CTTNBP2NL, SETD6, CACNB2, and UNC5C on the deregressed estimated breeding value for the SI traits days open (DO), number of inseminations per lactation (NI), success after first insemination (SFI), pregnancy within 70 d (P70), 90 d (P90), and 110 d (P110) after parity or for the conception rate (CR) (Table F).
References
- 1.Jamrozik J, Fatehi J, Kistemaker G J. (2005) Estimates of genetic parameters for Canadian Holstein female reproduction traits. , Journal of dairy science 88(6), 2199-2208.
- 2.González-Recio O, Alenda R. (2005) Genetic Parameters for female fertility traits and a fertility index in Spanish dairy cattle. , Journal of dairy science 88(9), 3282-3289.
- 3.Liu Z, Jaitner J, Reinhardt F. (2008) Genetic evaluation of fertility traits of dairy cattle using a multiple-trait animal model. , Journal of dairy science 91(11), 4333-4343.
- 4.Höglund J K, Guldbrandtsen B, Lund M S. (2015) Identification of genomic regions associated with female fertility in Danish Jersey using whole genome sequence data. , BMC genetics 16, 60.
- 5.Kawahara T, Gotoh Y, Suzuki M, Baba T, Yamaguchi S. (2012) Estimates of genetic parameters for female fertility traits and development of a fertility selection index in Japanese Holsteins. Proceedings of 15th AAAP Animal Science Congress,ThammasatUniversity,RangsitCampus,Bangkok/PathumThani,Thailand,26-30 Available: http://www.holstein.or.jp/hhac/kairyo/bindx/fullpaper_m0380-23_tkawahara.pdf .
- 6.Höglund J K, Buitenhuis B, Guldbrandtsen B. (2015) Genome-wide association study for female fertility in Nordic Red cattle. , BMC genetics 16, 110.
- 7.Minozzi G, E L Nicolazzi, Stella A. (2013) Genome wide analysis of fertility and production traits in Italian Holstein cattle. , PloS one; 8(11), 80219.
- 8.Sugimoto M, Sasaki S, Gotoh Y. (2013) Genetic variants related to gap junctions and hormone secretion influence conception rates in cows. Proceedings of the National Academy of Sciences of the United States of America 110(48), 19495-19500.
- 9.Kang H M, Sul J H, Service S K. (2010) Variance component model to account for sample structure in genome-wide association studies. , Nature 42(4), 348-354.
- 10.Zimin A V, Delcher A L, Florea L. (2009) A whole-genome assembly of the domestic cow, Bos taurus. Genome biology. 10(4), 42.
- 11.Porto-Neto L R, Kijas J W.Reverter A (2014)The extent of linkage disequilibrium in beef cattle breeds using high-density SNP genotypes. , Genetics, selection, evolution 46, 22.
- 12.Purcell S, Neale B, Todd-Brown K. (2007) PLINK: a toolset for whole-genome association and population-based linkage analysis. American journal of human genetics. 81(3), 559-575.
- 13.Sugimoto M, Gotoh Y, Kawahara T. (2015) Molecular effects of polymorphism in the 3’UTR of unc-5 homolog c associated with conception rate in holsteins. , PloS one; 10(7), 0131283.
- 14.Garrick D J, Taylor J F, Fernando R L. (2009) Deregressing estimated breeding values and weighting information for genomic regression analyses. , Genetics, selection, evolution 41, 55.
- 15.Niwa H, Yamamura K, Miyazaki J. (1991) Efficient selection for high-expression transfectants with a novel eukaryotic vector. , Gene 108(2), 193-199.
- 16.Xu Y, Morse L R, da Silva RA. (2010) PAMM: a redox regulatory protein that modulates osteoclast differentiation. Antioxidants & redox signaling. 13(1), 27-37.
- 17.Garnier J, Osguthorpe D J, Robson B. (1978) Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. , Journal of molecular biology 120(1), 97-120.
- 18.Agarwal A, Aponte-Mellado A, Premkumar B J. (2012) The effects of oxidative stress on female reproduction: a review. Reproductive biology and endocrinology. 10, 49.
- 19.Aurousseau B, Gruffat D, Durand D. (2006) Gestation linked radical oxygen species fluxes and vitamins and trace mineral deficiencies in the ruminant. , Reproduction, nutrition, development 46(6), 601-620.
- 20.Dixit V D, Parvizi N. (2001) Pregnancy stimulates secretion of adrenocorticotropin and nitric oxide from peripheral bovine lymphocytes. Biology of reproduction. 64(1), 242-248.
- 21.Yang C, Ling H, Zhang M. (2011) Oxidative stress mediates chemical hypoxia-induced injury and inflammation by activating NF-κb-COX-2 pathway in HaCaT cells. Molecules and cells. 31(6), 531-538.
- 22.Yang C, Yang Z, Zhang M. (2011) Hydrogen sulfide protects against chemical hypoxia-induced cytotoxicity and inflammation in HaCaT cells through inhibition of ROS/NF-κB/COX-2 pathway. PLoS one. 6(7), 21971.
- 23.Lim H, Paria B C, Das S K. (1997) Multiple female reproductive failures in cyclooxygenase 2-deficient mice. , Cell 91(2), 197-208.
- 24.Lim H, Gupta R A, Ma W G. (1999) Cyclo-oxygenase-2-derived prostacyclin mediates embryo implantation in the mouse via PPARdelta. , Genes & development 13(12), 1561-1574.
- 25.Ma L, Wiggans G R, Wang S. (2012) Effect of sample stratification on dairy GWAS results. , BMC genomics 13, 536.
- 26.Weller J I, Ron M. (2011) Invited review: Quantitative trait nucleotide determination in the era of genomic selection. , Journal of dairy science 94(3), 1082-1090.
- 27.Buttchereit N, Stamer E, Junge W. (2011) Short communication: genetic relationships among daily energy balance, feed intake, body condition score, and fat to protein ratio of milk in dairy cows. 94(3), 1586-1591.
- 28.Kozlov G, Määttänen P, Thomas D Y. (2010) A structural overview of the PDI family of proteins. The FEBS journal. 277(19), 3924-3936.
- 29.E1 Asselin, Lacroix D, Fortier M A. (1997) IFN-tau increases PGE2 production and COX-2 gene expression in the bovine endometrium in vitro.Molecularandcellularendocrinology;132(1-2):. 117-126.
- 30.Assal-Meliani A, Kinsky R, Martal J. (1995) In vivo immunosuppressive effects of recombinant ovine interferon-tau (trophoblastin): r.oTP (r.oIFN-tau) inhibits local GVH reaction in mice (PLN assay), prevents fetal resorptions, and favors embryo survival and implantation in the CBA/J x DBA/2 mice combination. American journal of reproductive immunology. 33(3), 267-275.
- 31.Chaouat G, Assal Meliani A, Martal J. (1995) IL-10 prevents naturally occurring fetal loss in the CBA x DBA/2 mating combination, and local defect in IL-10 production in this abortion-prone combination is corrected by in vivo injection of IFN-tau. , The Journal of immunology 154(9), 4261-4268.
- 32.Chen Q, Zhang W, Ran H. (2014) PKCδ and θ possibly mediate FSH-induced mouse oocyte maturation via NOX-ROS-TACE cascade signaling pathway. PLoS one. 9(10), 111423.
- 33.Shkolnik K, Tadmor A, Ben-Dor S. (2011) Reactive oxygen species are indispensable in ovulation. Proceedings of the National Academy of Sciences of the United States of America 108(4), 1462-1467.
Cited by (1)
- 1.Lei Qiuxia, Zhang Shuer, Wang Jie, Qi Chao, Liu Jie, et al, 2024, Genome-wide association studies of egg production traits by whole genome sequencing of Laiwu Black chicken, Poultry Science, 103(6), 103705, 10.1016/j.psj.2024.103705