Plasma TREM2 Levels, Alcohol Consumption, and Liver Enzymes in Patients with Alcohol use Disorder: A Sex-Dependent Relationship Involving MS4A6A Genetic Polymorphism
Abstract
Alcohol use disorder (AUD) is the most prevalent substance use disorder. Excessive alcohol consumption leads to a range of health issues. We set out to identify inflammatory markers linked to alcohol consumption, which might ultimately offer novel insight into genetic underpinnings and have implications for alcohol-associated disease. Alcohol consumption and blood-based multi-omics data were collected by The Mayo Clinic Center for Individualized Treatment of Alcohol Dependence study. Plasma samples from patients with AUD were used for proteomics analysis using the OLINK “Explore Inflammation” panel (n=410). Liver enzymes were also measured. A genome-wide association study (GWAS) was performed to explore the relationship between genetic variants and plasma TREM2 levels. Our findings show thatplasma triggering receptor expressed on myeloid cells 2 (TREM2), a key gene associated with neurodegenerative disease, was the most significant signal correlated with alcohol consumption, and has also been associated with liver enzyme levels in patients with AUD. We identified the rs7232 single nucleotide polymorphism (SNP) in MS4A6A as a key genetic variant associated with plasma TREM2 levels, with the minor allele (A) linked to higher TREM2 levels and increased alcohol consumption, particularly in men. Furthermore, MA4A6A is an ethanol-responsive gene in a SNP-dependent manner, and the variant genotype of the rs7232 SNP was associated with lower expression for MA4A6A due to proteasome-mediated protein degradation. In summary, this study provides insight into the relationship between plasma TREM2 levels, alcohol consumption, and liver function in AUD patients, shedding light on genetic factors underlying alcohol-related diseases.
Author Contributions
Copyright © 2025 Ming-Fen Ho, et al
 This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Competing interests
Dr Weinshilboum is a cofounder of and stockholder in OneOme LLC, a pharmacogenomics decision -support company. Dr. Croarkin has received research grant support from Neuronetics, NeoSync and Pfizer, Inc. He has also received grants-in-kind (equipment support for research studies) from Assurex; MagVenture, Inc; and Neuronetics, Inc. Dr. Croarkin has also served on advisory boards for Engrail Therapeutics, Myriad Neuroscience, Procter & Gamble, and Sunovion. All other authors have no conflicts to declare.
Citation:
Introduction
Alcohol use disorder (AUD) is the most prevalent substance use disorder (SUD) worldwide 1, 2. Excessive alcohol use can impact bodily systems, raising the risk of liver disease, neurodegenerative disease, mental disorders, and more 3, 4. An extensive literature suggests that genetic predisposition to alcohol consumption is significantly correlated with disease risk, including chronic pancreatitis, cancer, and cardiovascular disease 5, 6, 7. Additionally, it has been reported that AUD is a major risk factor for the onset of multiple types of dementia, especially early onset dementia 8. However, to date there are no biomarker for alcohol-associated disease. Recent discoveries in the UK biobank showed that utilizing multi-omics strategies could enhance the signals for a large number of known gene-disease associations, indicating that well-defined clinical phenotypes coupled with biological data may be useful to uncover disease-risk biomarkers 9. We therefore set out to identify biomarkers associated with alcohol consumption, ultimately providing novel insight into genetic underpinnings and their implications for alcohol-associated diseases.
Currently, the most common assessment tools for alcohol consumption include the alcohol use disorders identification test (AUDIT), and timeline follow-back (TLFB), all of which are self-report of quantity and frequency of drinking. In addition, several laboratory tests have been used to quantify an individual’s alcohol consumption, and many of those tests are correlated with levels of specific liver enzymes, including aspartate aminotransferase (AST), alanine aminotransferase (ALT), and gamma-glutamyl transferase (GGT). Those enzymes are useful indicators of liver function, however, they are not biomarkers for specific alcohol-related disease with the exception of alcohol-associated liver disease 10.
The Mayo Clinic Center for Individualized Treatment of Alcohol Dependence study contains dense clinical information, including alcohol consumption and blood-based multi-omics data 11, 12. Plasma proteomics was performed using the Olink Explore 384 Inflammation panel, which is currently the most comprehensive and most sensitive high-performance protein biomarker panel available for inflammation studies 13, 14, 15. We previously used these sets of data to identify potential inflammatory markers associated with acamprosate treatment outcomes 12, 16. These population-level multi-omics datasets offer the opportunity to discover novel gene-disease biomarkers and to explore their biological connections 12, 17, 18. Given that chronic alcohol consumption is a risk factor for many chronic diseases and conditions, this study was designed to identify plasma protein markers associated with recent alcohol consumption using our established proteomics-informed genomics approach. These findings may provide insight into genetic underpinnings together with their implications for alcohol associated diseases.
Our proteomics-informed genome-wide association study (GWAS) research approach, as illustrated in Figure 1, was designed to integrate data from proteomics with genomic information to obtain a deeper understanding of biological mechanisms 12, 17, 18. By combining proteomics and genomics, we can pinpoint genetic variation that affects protein concentrations, identify biomarkers and understand underlying biological mechanisms. This approach has been used in individualized medicine, drug discovery and understanding molecular mechanisms underlying various neuropsychiatric disorders. Our research team has successfully applied this approach to identify biomarkers for drug treatment response, i.e. acamprosate for AUD 12, 17, and selective serotonin reuptake inhibitors (SSRIs) for major depressive disorder 19. As a first step, we determined whether proteomics profiles were associated with recent alcohol use. We then performed GWAS to identify candidate biomarkers that might contribute to alcohol use and alcohol-related diseases. We subsequently investigated whether any single nucleotide polymorphisms (SNPs) identified in our GWAS might also be associated with alcohol-related disease and then explored underlying molecular mechanisms (Figure 1).
Figure 1.Proteomics-inform genomics research strategy.
Our results highlighted plasma triggering receptor expressed on myeloid cells 2 (TREM2) as a significant biomarker associated with recent alcohol consumption, showing robust correlations with various alcohol consumption metrics, including the total number of drinks, drinking days, and heavy drinking days. TREM2 also plays a role in immune response, particularly in microglial function and inflammatory processes in the brain 20, 21. Furthermore, this gene has significant implications for neurodegenerative diseases such as Alzheimer’s disease. Moreover, the positive associations between plasma levels of TREM2 and liver enzymes, such as GGT, AST, and ALT, suggest that TREM2 may be linked to alcohol intake as well as the liver’s response to chronic alcohol consumption. Finally, we performed functional genomic studies to demonstrate new biological insights into the relationship between plasma TREM2 levels, genetic variation, alcohol consumption, liver function and alcohol-associated disease in patients with AUD.
Methods and materials
Study participants and ethics statements
We previously recruited individuals with AUD for the Mayo Clinic Center for the Individualized Treatment of Alcohol Dependence Study (ClinicalTrials.gov Identifier: NCT00662571). All subjects whose biological samples were included in this study provided their consent to participate in the study and publish the study results in peer reviewed journals. This study was conducted under the protocol reviewed and approved by the Mayo Clinic Institutional Review Board (reference IRB number: 18-006428). We used TLFB to collect alcohol use information for 90 days before blood collection. The sample size was based on plasma sample availability (n=410).
Targeted proteomics
The OLINK “Explore Inflammation” panel was assayed using EDTA-plasma samples (n=410) 12. We assayed all plasma samples in a single batch. Briefly, plasma samples (1µL) from each subject were incubated in the presence of proximity antibody pairs linked to DNA reporter molecules. Data were normalized to standard EDTA plasma controls to produce relative protein abundance information using normalized protein expression (NPX) manager software, which determined protein concentration on a log2 scale. The average intra-assay and inter-assay coefficients of variation (CV) were 9% - 14% across the five plates studied. Liver enzymes including gamma-glutamyl transferase (GGT), aspartate aminotransferase (AST), and alanine aminotransferase (ALT) were also measured in the Mayo research core facility 17.
GWAS for plasma TREM2 levels
DNA samples were initially genotyped using Illumina Human Core arrays in the Mayo Clinic Medical Genome Facility. The National Institute of Alcohol Abuse and Alcoholism then genotyped these samples using Infinium OmniExpressExome-8 BeadChips. Data from the two arrays were quality-controlled, combined and checked for concordance and additional quality control was performed using the combined dataset 22. Some samples were excluded from analysis because of low call rate, extreme heterozygosity, or disagreement between reported sex and genetically determined sex. Sample relatedness was checked by pairwise identical-by-descent estimation. Imputation was conducted using the Michigan Imputation Server with the haplotype reference consortium (HRC) reference panel (version HRC.r1-1.GRCh37.wgs.mac5.sites). SNPs with a call rate <95%, SNPs not in Hardy -Weinberg equilibrium and SNPs with minor allele frequency (MAF) <0.01 among the patients with AUD were excluded. Of the 6,654,675 SNPs studied, 6,621,773 passed initial quality control and were used in the genome-wide association study (GWAS). Associations between variants and plasma TREM2 levels were examined using linear regression, adjusted for sex, age, and study site. These analyses were done in R version 4.4.1 and PLINK 1.9.
Site-directed mutagenesis and protein expression quantification
A human MS4A6A cDNA clone (OHu27116) was purchased from GenScript (New Jersey, USA). This cDNA clone was used as template for the construction of mutant plasmid (T185S). The sequences of all constructs were verified by DNA sequencing and proved that only the expected mutation had occurred. HEK293T cells were transfected using Lipofectamine 3000 (Invitrogen) with 0.5 μg of the plasmid DNA. Expression was quantified by Western Blot. Briefly, protein samples were quantified using BCA protein assay kits (Thermo Scientific, cat#: 23227). Protein (10 μg) samples were loaded on 4-20 % gradient SDS-PAGE precast Gels and transferred onto polyvinylidene fluoride membranes. After blocking, membranes were incubated with primary antibodies against MS4A6A or beta-actin at 4°C overnight. The washed membranes were then incubated with secondary antibody for an hour at room temperature. The membranes were then incubated in ECL substrate (Thermo Scientific, Madison, WI, USA) for one minute, and were visualized with the Geldoc Go Gel imaging system (Bio-Rad, USA). Protein expression levels were quantified using Bio-Rad image lab software. For some experiments, cells were treated with 10μM of MG-132, a potent, reversible, and cell-permeable proteasome inhibitor (MedChemExpress, cat: HY-13259) or 3-methyladenine (3-MA), a reagent that has been widely used to block autophagy (MedChemExpress, cat: HY- 19312) for 8 hours, and 24 hours respectively. Cell lysates were then collected for Western Blot analysis. Three independent experiments were performed.
Statistical analysis
Statistical analyses were performed using R Statistical Software (version 4.4.1), and figures were generated using GraphPad Prism Software v7. Experiments were performed using randomization and blinding analysis when technically feasible and appropriate. Log-transformed proteomics data were analyzed. Gene expression was analyzed using analysis of variance (ANOVA), followed by planed comparisons with appropriate post hoc tests. We also conducted t-tests for all meaningful combinations (effect of treatments on wildtype and variant, effect of genotype on treatments vs vehicle). The exact sample size for each experiment is listed in the figure legends.
Data availability
All data supporting our findings can be found in the main paper or in supplementary files.
Results
Plasma TREM2 levels associated with recent alcohol use and liver enzyme concentrations
Among the AUD patients included in this study, approximately two-thirds were men, with an average age for all participants of 41.99 ± 11.74 years at the time of consent (Table 1). The average age of onset for AUD was 29.00 ± 11.91 years, indicating a long history of drinking among the patients. This study utilized TLFB, a self-report alcohol consumption measure, to evaluate daily consumption of drinks within the past 90 days. The 2020-2025 U.S. Dietary Guidelines recommend that moderate drinking be limited to one drink or less per day for women and two drinks or less per day for men. We found that the men in our study drank more than the women (Table 1). Specifically, the average number of drinks per day in the past three months was 9.1 for women and 13.1 for men (Table 1).
Table 1. Clinical characteristics of the study cohort.| Men + Women (n=410) | Men (n=267) | Women (n=143) | Men vs Women | ||||
| Mean | Std | Mean | Std | Mean | Std | P value* | |
| AUD subjects | |||||||
| Age | 41.99 | 11.74 | 42.2 | 11.72 | 41.6 | 11.8 | 0.6201 |
| Age of onset for AUD | 29 | 11.91 | 28.37 | 11.92 | 30.16 | 11.86 | 0.155 |
| Total drinks+ | 562.8 | 513.2 | 630.9 | 538.5 | 434.8 | 435.7 | 0.0002 |
| Number of drinking days+ | 45.36 | 27.23 | 46.12 | 27.59 | 43.93 | 26.58 | 0.4402 |
| Number of heavy drinking days+ | 41.48 | 27.16 | 42.62 | 27.71 | 39.35 | 26.06 | 0.2459 |
| Average drinks per day+ | 11.85 | 7.905 | 13.13 | 8.419 | 9.441 | 6.177 | <0.0001 |
| Average drinks per week+ | 43.66 | 40 | 49.03 | 42.09 | 33.56 | 33.64 | 0.0002 |
| Average drinks per months+ | 187.1 | 171.4 | 210.1 | 180.4 | 143.8 | 144.2 | 0.0002 |
| GGT (U/L) | 69.41 | 98.25 | 79.38 | 101 | 49.02 | 75.36 | 0.0046 |
| AST (U/L) | 38.2 | 34.65 | 38.65 | 34.3 | 36.67 | 36.37 | 0.6547 |
| ALT (U/L) | 46.31 | 56.93 | 53.04 | 68.8 | 33.72 | 25.37 | 0.0398 |
Chronic alcohol consumption is a risk factor for many chronic diseases and conditions 23, 24. We designed this study to identify plasma biomarkers associated with alcohol consumption which might provide insight into the genetic underpinnings together with its implications for alcohol associated diseases. We found that plasma TREM2 was the most significant plasma protein marker positively correlated with recent alcohol consumption metrics, including the total number of drinks (r: 0.31, p: 1.15E-10), number of drinking days (r: 0.29, p: 2.45E-09), and number of heavy drinking days (r: 0.30, p: 1.03E-09) as determined by TLFB (Figure 2A, for complete data see Supplementary Tables 2-4). Additionally, GGT, a marker for recent heavy alcohol use, also showed a positive correlation with plasma TREM2 concentration (r: 0.36, p: 1.52E-11) (Figure 2B). In line with these observations, plasma TREM2 levels were positively correlated with liver enzymes, specifically AST (r: 0.32, p: 4.95E-08) and ALT (r: 0.26, p: 1.13E-05), suggesting a link between plasma TREM2 levels, liver function and alcohol consumption (Figure 2B).
Figure 2.Plasma TREM2 may be a biomarker for recent alcohol use. Plasma TREM2 levels positively correlated with (A) recent alcohol consumption and (B) concentrations of liver enzymes in patients with AUD.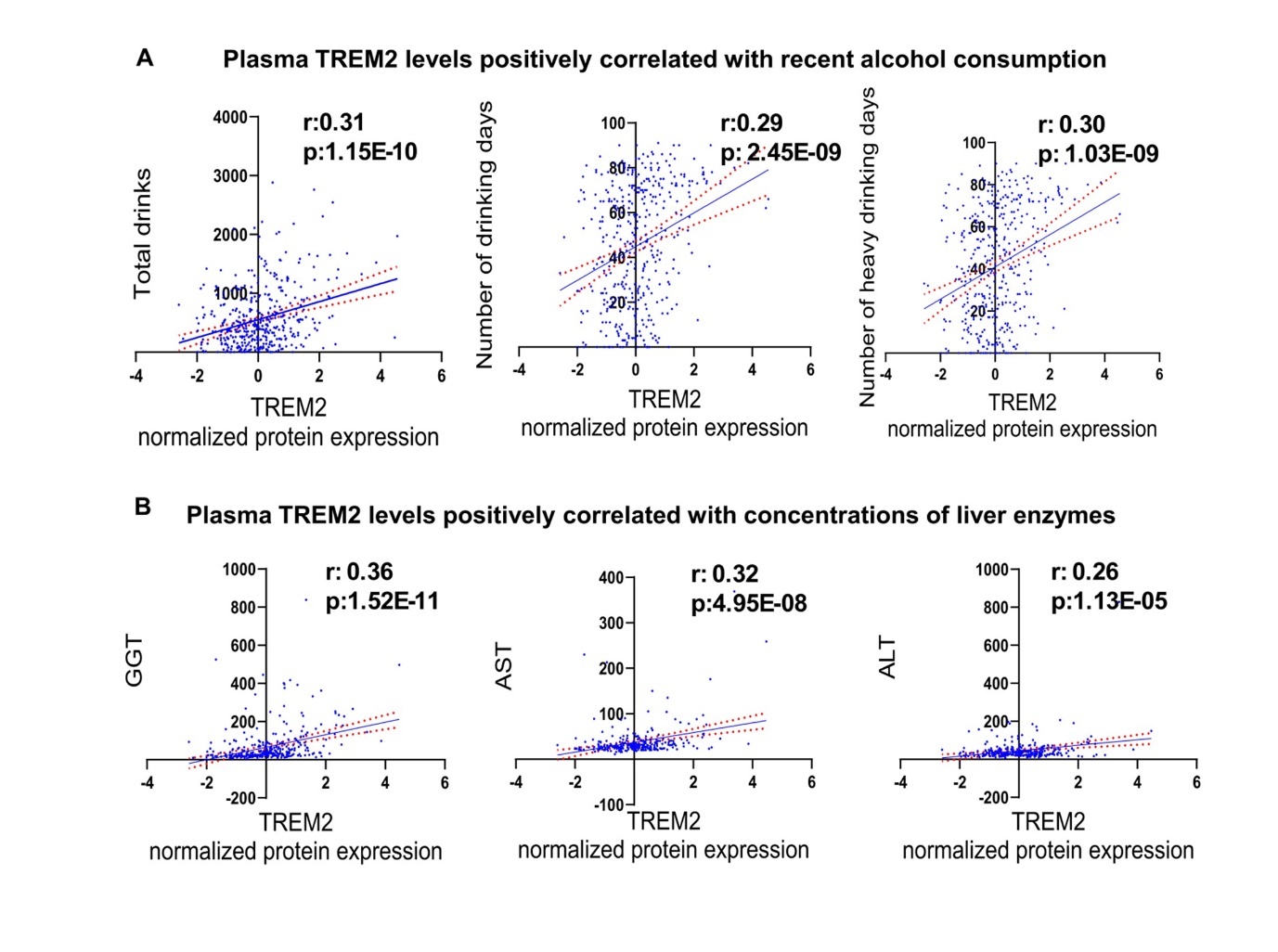
GWAS for plasma TREM2 concentrations
The relationship between alcohol use and TREM2 expression remains unclear. Thus, we applied our "proteomics-informed GWAS" approach to identify potential genes and genetic variants influencing plasma TREM2 levels (Figure 1). The GWAS for plasma TREM2 concentrations revealed a genome-wide significant signal (rs140498820, p: 2.09E-09) on chromosome 19, which mapped to ADGRE4P (Adhesion G protein-coupled receptor E4), a pseudogene (Figure 3A, and Supplementary Table 4). Notably, rs7232, a missense variant in MS4A6A (Membrane-spanning 4-domains subfamily A member 6A), emerged as one of the top signals (p: 2.59E-07) (Figure 3A- Figure 3B), which we found is functional and may have implications for alcohol consumption, as described in subsequent paragraphs.
Figure 3.GWAS for plasma TREM2 levels in patients with AUD. (A) Manhattan plot for GWAS of plasma levels of TREM2. (B) Locus zoom plot for the rs7232 SNP, which mapped to MS4A6A on chromosome 11. The rs7232 SNP was associated with alcohol consumption in a SNP and sex-dependent fashion. Patients with the A allele of the rs7232 SNP reported (C) more drinks, and (D) more drinking days in the past month.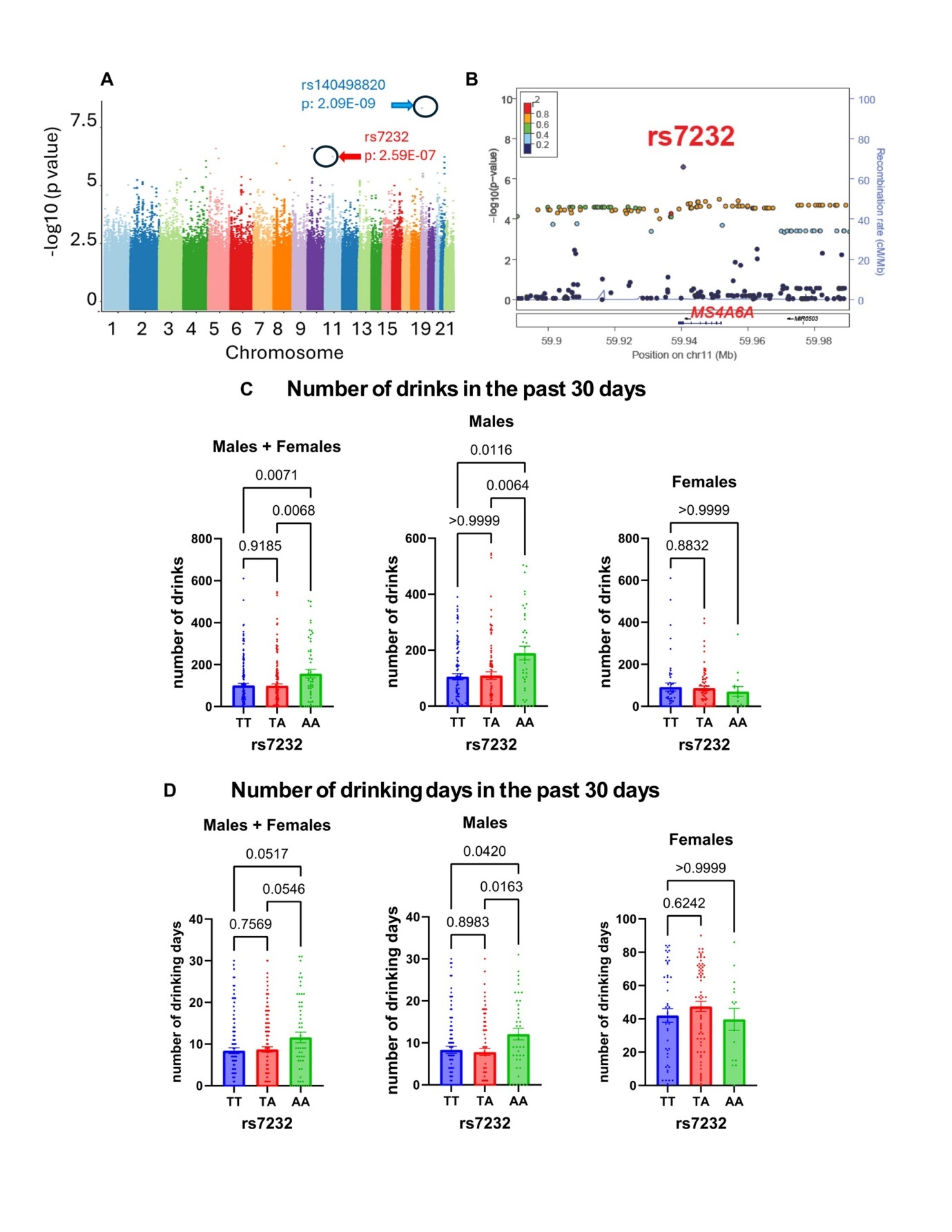
A SNP-dependent relationship between the MS4A6A genetic polymorphism and alcohol consumption
Patients with the AA genotype for rs7232 reported higher alcohol consumption in the past month (Figure 3C). Subsequently, we analyzed our data by sex and discovered that the AA genotype for rs7232 was associated with an increase in alcohol consumption, i.e. number of drinks and number of drinking days in the past one month, among male subjects (Figure 3D). However, we did not observe a SNP-dependent pattern of alcohol consumption in women, which may be at least partially due to small sample size. As mentioned previously, two thirds of the study participants were men (Table 1). The rs7232 SNP has been associated previously with plasma TREM2 concentrations (p: 3.0E-190, n= 10708) 25. Consistent with our findings, the minor allele (A) was associated with higher plasma TREM2 levels (Figure 4A). The minor allele frequency for rs7232 is 37% in European Americans. The minor allele (A) is associated with lower MS4A6A expression (p: 1.44E-12) in whole blood, based on the GTEx database (Figure 4B). Furthermore, MS4A6A is an ethanol-responsive gene but this response is only observed in cells transfected with the wildtype SNP. Specifically, cells transfected with the wildtype plasmid revealed a significantly higher level of MS4A6A compared to those transfected with the variant genotype in the absence of ethanol. Upon ethanol treatment, the mRNA level of MS4A6A decreased in cells transfected with the wildtype plasmid, whereas no change was observed in the mRNA level of MS4A6A after ethanol exposure in cells transfected with the variant plasmid (Figure 4C).
MS4A6A missense variant caused proteasome-mediated protein degradation
This genetic polymorphism (rs7232) substituted amino acid threonine to serine, and is predicted to be “possibly damaging”, with a score of 0.827 (sensitivity: 0.84; specificity: 0.93) by prediction of the functional effects of human non-synonymous single nucleotide polymorphisms software (Poly Phen-2 software) 26, and Combined Annotation Dependent Depletion (CADD v1.7) also indicated that this genetic polymorphism might be deleterious 27. As the rs7232 SNP was an eQTL for the MS4A6A gene (Figure 4B), we tested the possibility that this missense variant might affect MS4A6A protein expression. MS4A6A plasmids that were wildtype or contained the nonsynonymous SNP (rs7232) were expressed in HEK-293T cell. We observed that rs7232, a SNP that encoded a T185S substitution, resulted in a significant decrease in MS4A6A protein expression (65±12 % of wildtype, p < 0.05) (Figure 4D). We next set out to determine the mechanism underlying the SNP-dependent protein degradation.
Proteasome-mediated degradation is a common functional mechanism for the effect of missense variants 19, 28, 29. Accelerated protein degradation is also a possible mechanism for the decreased level of protein displayed by the variant genotype, which could be due to ubiquitin-proteasome or autophagy-mediated degradation. To investigate these possibilities, we set out to test the effects of a protease inhibitor (MG-132) or an autophagy inhibitor (3-MA) on MS4A6A protein expression in HEK-293T cells that were transfected with plasmids encoding MS4A6A that was wildtype or contained one amino acid substitution (T185S). Our results showed that the protease inhibitor, MG-132, increased the protein level of MS4A6A in cells with the variant genotype (Figure 4E), but the autophagy inhibitor (3-MA) did not have any effect, indicating that the degradation of MS4A6A protein was proteasome -mediated (Figure 4F).
Taken together, our findings indicate a positive correlation between plasma TREM2 levels and recent alcohol consumption. Through a proteomics-informed GWAS approach, we identified the rs7232 SNP, a missense variant in MS4A6A, as a potential key genetic factor influencing plasma TREM2 levels. The minor allele (A) was associated with increased plasma TREM2 levels and reduced MS4A6A expression. Furthermore, our functional genomic studies demonstrated that the SNP-dependent gene expression could be linked to proteasome-mediated protein degradation.
Figure 4.The rs7232 SNP is functional. (A) SNP-dependent plasma TREM2 concentrations. (B) The rs7232 SNP is an eQTL for MS4A6A in whole blood based on the GTEx database. (C) we performed functional genomic studies to demonstrate that MA4A6A is an ethanol-responsive gene in a SNP-dependent manner. (D) MS4A6A plasmids that were wildtype or contained the nonsynonymous SNP (rs7232) were expressed in HEK-293T cell. The rs7232 SNP that encoded a T185S substitution, resulted in a significant decrease in MS4A6A protein expression. Protein expression was determined by Western blot analysis. Protein quantification is shown in the bottom panel. A Student’s t test was performed to compare gene expression in cells with differing MS4A6A SNP genotypes (wildtype vs variant). *A p value ≤0.05 was considered statistically significant. All values are mean +/- S.E.M for three separate independent assays. (E) MG-132 increased the protein expression of MS4A6A in cells that expressed the variant genotype for the rs7232 SNP. (F) 3-MA did not have effects on the protein expression of MS4A6A. Three independent experiments were performed. Values are mean ± SEM of three assays.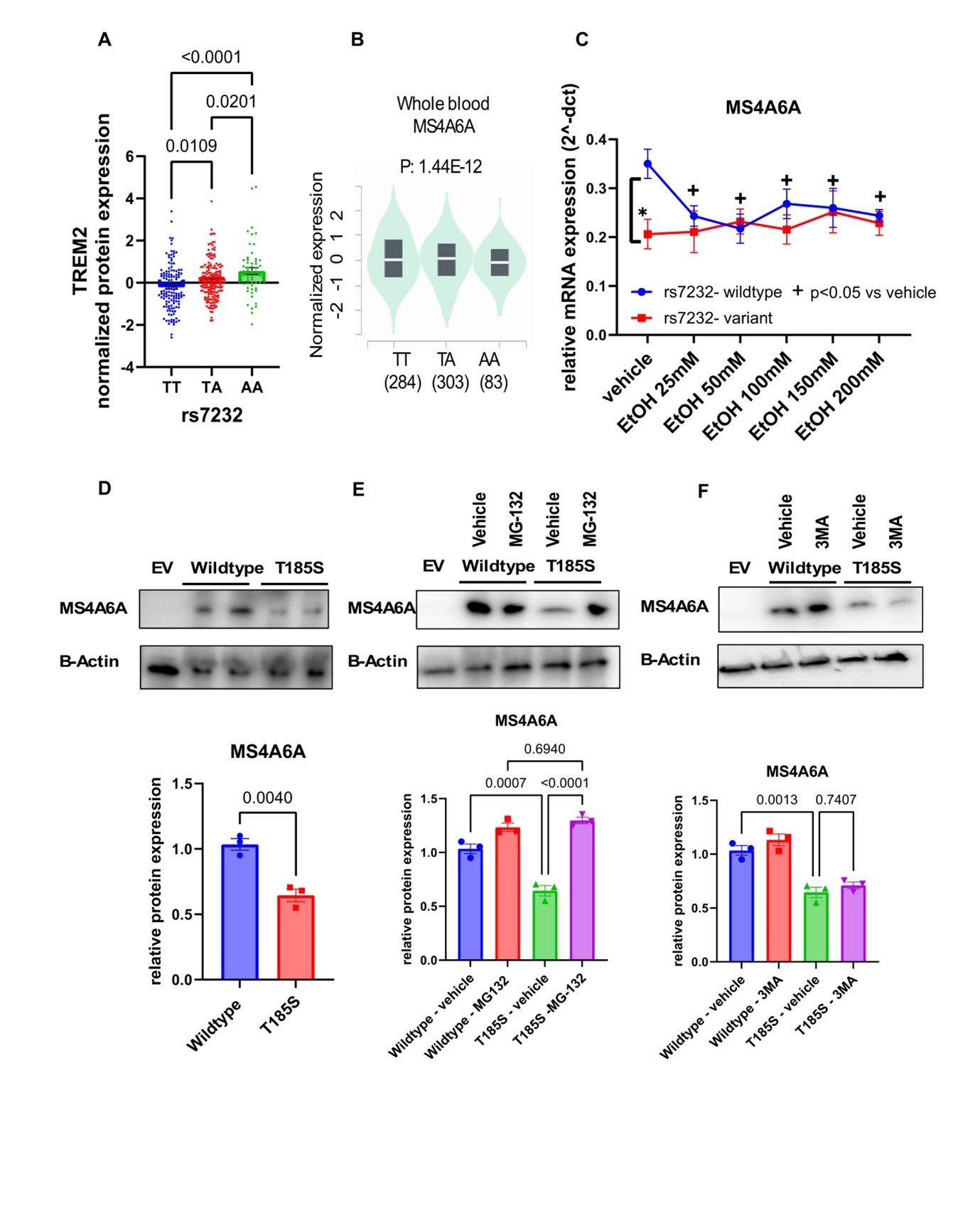
Discussion
The Mayo Clinic Center for Individualized Treatment of Alcohol Dependence study is an open-label acamprosate clinical trial which was originally designed to identify biomarkers associated with acamprosate treatment response 12, 17. In the present study, we set out to leverage the existing multi-omics data from that trial to identify biomarkers that might provide insight into the genetic underpinnings of alcohol consumption and its broader implications for alcohol associated diseases.
Our analysis revealed plasma TREM2 as a key marker associated with recent alcohol consumption, showing robust correlations with various self-report alcohol consumption metrics, including the total number of drinks, drinking days, and heavy drinking days. Moreover, we observed significant positive associations between plasma TREM2 levels and levels of liver enzymes, such as GGT, AST, and ALT. While we could not establish causality, the significant correlation between levels of plasma TREM2 and liver enzymes implies that TREM2 might serve as an indicator of alcohol-induced liver injury, a critical aspect of AUD pathophysiology. Given that plasma TREM2 has previously been proposed as a circulating marker of non-alcoholic fatty liver disease 30, its involvement in systemic inflammation and its potential link to alcohol-induced liver damage add a new dimension to our understanding of TREM2's role in AUD. TREM2 is also known for its role in the immune system, particularly in microglial function and inflammatory processes in the brain 31, 32, 33. Its link to systemic inflammation, particularly in the context of chronic alcohol use, underscores the broader implications of TREM2 dysregulation in AUD.
The identification of TREM2 as a biomarker not only provides a tool for assessing alcohol consumption and its health impacts but also aligns with the ongoing effort to bridge the gap between psychiatric phenotypes and their biological underpinnings. One of the major challenges in psychiatric research is the limited connection between “well-defined” clinical phenotypes and the underlying biology. This is especially true for conditions such as alcohol addiction, where the lack of biological measures impedes diagnostic precision, and limits the ability to monitor disease progression. To address this challenge, we applied our established “proteomics-informed genomic” approach, integrating biological quantitative measures with clinical traits i.e. alcohol use, to identify relevant biomarkers. Our findings highlighted plasma TREM2 as a biomarker for recent alcohol consumption and highlighted the role of genetic variation, particularly the rs7232 SNP in the MS4A6A gene, modulating its levels in patients with AUD (Figure 3).
Our proteomics-informed GWAS identified the rs7232 SNP in MS4A6A as significantly associated with plasma TREM2 levels (Figure 3). This aligns with earlier studies that have linked the MS4A gene family to TREM2 regulation, particularly in the context of neuroinflammation and Alzheimer’s disease 34, 35. Our study extends these associations to AUD, suggesting that genetic variation in MS4A6A may influence TREM2 expression and, consequently, the inflammatory response to alcohol consumption. The rs7232 SNP is associated with both increased plasma TREM2 levels and decreased MS4A6A protein expression, likely through proteasome-mediated degradation (Figure 4). These observations provide possible mechanistic explanation for the SNP's biological effects. Notably, we found that the association between the rs7232 SNP and alcohol consumption was more pronounced in men than in women. This sex-dependent difference could be due to the larger proportion of male participants in our study, but it also raises questions about how genetic factors might differentially affect the risk of alcohol-related diseases in men and women. Further research with a more balanced sample of male and female participants is needed to understand the sex-specific genetic influence on alcohol consumption and TREM2 levels.
While the present study provides important insights, it is not without limitations. First, the sample size, particularly the smaller number of female participants, may limit the generalizability of our findings across sexes. Second, while we identified a significant genetic association with plasma TREM2 levels, the exact mechanisms by which alcohol exposure influences TREM2 expression and its downstream effects on liver function require further investigation. Future studies are needed to replicate our findings in larger, more diverse cohorts and to explore the longitudinal relationships between TREM2, alcohol consumption, and alcohol-related diseases. Nevertheless, this series of analyses enhanced our understanding of the molecular and genetic underpinnings of AUD and suggested new avenues for research into biomarkers for alcohol-related diseases.
It is well-documented that TREM2 is a known risk gene for Alzheimer's disease (AD) 36, 37. It is important to note that the rs7232 SNP has previously been associated with TREM2 concentrations in both plasma and cerebrospinal fluid (CSF). Pietzner et al. reported that rs7232 is linked with plasma TREM2 (p: 3.0E-190, n= 10708) 25 Moreover, it has been reported that rs7232 was associated with CSF soluble TREM2 levels in two independent studies, including an Alzheimer’s Disease Neuroimaging Initiative (ADNI) study cohort (p: 1 x 10-14, n=1001) 34, and a Chinese Alzheimer's Biomarker and LifestylE study (p = 1.42 × 10-15, n=449) 35. Given the association of rs7232 with TREM2 concentrations and Alzheimer's disease risk, these findings highlight the complex interplay between TREM2 expression, alcohol use, and alcohol-related neurodegenerative diseases. The shared pathways involving TREM2 in both AUD and Alzheimer’s disease suggest that further investigation into this gene’s role could provide critical insights into the overlapping mechanisms of alcohol-related liver damage and neuroinflammation. This series of analyses enhances our understanding of TREM2’s role in AUD and highlights its potential as a biomarker for both alcohol consumption and alcohol-induced liver injury. These findings open new avenues for future research into the genetic regulation of TREM2 and its implications for alcohol-related diseases, with potential to inform personalized interventions for patients suffering from AUD.
Conclusions
This series of experiments could potentially advance our understanding of the genetic and molecular mechanisms underlying the regulation of plasma TREM2 levels. TREM 2 is also ethanol inducible, which suggests its implications in alcohol-associated diseases. This study sheds light on how MS4A6A genetic variation (rs7232) can influence TREM2 levels. Additionally, the link between this SNP and proteasome-mediated protein degradation offers insight into the broader biological pathways that may be involved. These findings could inform future research on therapeutic targets and biomarkers for alcohol-related diseases in which TREM2 plays a crucial role.
Funding
This work was supported in part by National Institutes of Health (grant numbers P20 AA17830, K01 AA28050, R01 AA27486 and R01 DA57928); the Brain & Behavior Research Foundation (grant number: 31329), the Mayo Clinic Research Pipeline K2R Program, Donna Giordano, the Terrance and Bette Noble Foundation and the Mayo Clinic Center for Individualized Medicine.
Author contributions
MH wrote the first draft of the manuscript and supervised the study. MH designed the research. MH, and CZ performed the research. MH, CZ, JB, and BC analyzed the data. CZ, BC, JB, and HL contributed analytical tools. TO, PC, and VK recruited study subjects. MS provided operational support. MH and RM obtained the funding. All authors have given final approval of the version to be published.
Supplementary Material
Acknowledgements
We extend our sincere gratitude to Dr. Leila Jones for providing administrative support for our addiction research program.
Abbreviations
References
- 1.Tucker J A, Chandler S D, Witkiewitz K. (2020) Epidemiology of Recovery From Alcohol Use Disorder. Alcohol Res. 40-3.
- 2.Suen L W, Makam A N, Snyder H R, Repplinger D, Kushel M B et al. (2021) . National Prevalence of Alcohol and Other Substance Use Disorders Among Emergency Department Visits and Hospitalizations: NHAMCS 2014-2018. J Gen Intern Med 1-9.
- 3.Abbas D, Ciricillo J A, Elom H A, Moon A M. (2023) Extrahepatic Health Effects of Alcohol Use and Alcohol-associated Liver Disease. Clinical Therapeutics. 45(12), 1201-1211.
- 4.Kamal H, Tan G C, Ibrahim S F, Shaikh M F, Mohamed I N et al. (2020) Alcohol Use Disorder, Neurodegeneration, Alzheimer’s and Parkinson’s Disease: Interplay Between Oxidative Stress, Neuroimmune Response and Excitotoxicity. Frontiers in Cellular Neuroscience. 14.
- 5.Li M, Zhang X, Chen K, Miao Y, Xu Y et al. (2024) . Alcohol Exposure and Disease Associations: A Mendelian Randomization and Meta-Analysis on Weekly Consumption and Problematic Drinking. Nutrients 16(10), 1517.
- 6.Biddinger K J, Emdin C A, Haas M E, Wang M, Hindy G et al. (2022) . , Association of Habitual Alcohol Intake With Risk of Cardiovascular Disease. JAMA Network Open 5(3), 223849-223849.
- 7.Halford J L, Weng L-C, Choi S H, Jurgens S J, Morrill V N et al. (2020) Associations Between Alcohol Intake and Genetic Predisposition With Atrial Fibrillation Risk in a National Biobank. Circulation: Genomic and Precision Medicine. 13-6.
- 8.Schwarzinger M, Pollock B G, OSM Hasan, Dufouil C, Rehm J et al. (2018) Contribution of alcohol use disorders to the burden of dementia in France 2008–13: a nationwide retrospective cohort study. The Lancet Public Health. 3-3.
- 9.Garg M, Karpinski M, Matelska D, Middleton L, Burren O S et al. (2024) Disease prediction with multi-omics and biomarkers empowers case–control genetic discoveries in the UK Biobank. Nature Genetics. 56(9), 1821-1831.
- 10.Kalas M A, Chavez L, Leon M, Taweesedt P T, Surani S. (2021) Abnormal liver enzymes: A review for clinicians. 13(11), 1688-1698.
- 11.Karpyak V M, Geske J R, Hall-Flavin D K, Loukianova L L, Schneekloth T D et al. (2019) Sex-specific association of depressive disorder and transient emotional states with alcohol consumption in male and female alcoholics. Drug and Alcohol Dependence. 196-31.
- 12.Ho M-F, Zhang C, Cohan J S, Tuncturk M, Heider R M et al. (2024) IL17RB genetic variants are associated with acamprosate treatment response in patients with alcohol use disorder: A proteomics-informed genomics. 120-304.
- 13.Agasing A M, Wu Q, Khatri B, Borisow N, Ruprecht K et al. (2020) Transcriptomics and proteomics reveal a cooperation between interferon and T-helper 17 cells in neuromyelitis optica. Nature Communications. 11(1), 2856.
- 14.Arunachalam P S, Wimmers F, CKP Mok, RAPM Perera, Scott M et al. (2020) Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. , Science 369(6508), 1210.
- 15.Hillary R F, Trejo-Banos D, Kousathanas A, McCartney D L, Harris S E et al. (2020) Multi-method genome- and epigenome-wide studies of inflammatory protein levels in healthy older adults. Genome Medicine. 12-1.
- 16.Ho M-F, Zhang C, Moon I, Coombes B J, Biernacka J et al. (2022) Plasma TNFSF10 levels associated with acamprosate treatment response in patients with alcohol use disorder. Frontiers in Pharmacology. 13-986238.
- 17.Ho M F, Zhang C, Moon I, Wei L, Coombes B et al. (2022) Genome-wide association study for circulating FGF21 in patients with alcohol use disorder: molecular links between the SNHG16 locus and catecholamine metabolism. Molecular Metabolism. 63, 101534.
- 18.Ho M-F, Zhang C, Wei L, Zhang L, Moon I et al. (2022) Genetic variants associated with acamprosate treatment response in alcohol use disorder patients: A multiple omics study. British journal of pharmacology. 173-13.
- 19.Gupta M, Neavin D, Liu D, Biernacka J, Hall-Flavin D et al. (2016) TSPAN5, ERICH3 and selective serotonin reuptake inhibitors in major depressive disorder: pharmacometabolomics-informed pharmacogenomics. Molecular psychiatry. 21(12), 1717-1725.
- 20.Liu W, Taso O, Wang R, Bayram S, Graham A C et al. (2020) Trem2 promotes anti-inflammatory responses in microglia and is suppressed under pro-inflammatory conditions. Hum Mol Genet. 29(19), 3224-3248.
- 22.Biernacka J M, Coombes B J, Batzler A, AM-C Ho, Geske J R et al. (2021) Genetic contributions to alcohol use disorder treatment outcomes: a genome-wide pharmacogenomics study. , Neuropsychopharmacology 46(12), 2132-2139.
- 23.Shield K D, Parry C, Rehm J. (2013) Chronic diseases and conditions related to alcohol use. Alcohol Res. 35(2), 155-173.
- 24.Sterling S A, Palzes V A, Lu Y, Kline-Simon A H, Parthasarathy S et al. (2020) Associations Between Medical Conditions and Alcohol Consumption Levels in an Adult Primary Care Population. JAMA Network Open. 3-5.
- 25.Pietzner M, Wheeler E, Carrasco-Zanini J, Cortes A, Koprulu M et al. (2021) Mapping the proteo-genomic convergence of human diseases. , Science 374(6569), 1541.
- 26.Adzhubei I A, Schmidt S, Peshkin L, Ramensky V E, Gerasimova A et al. (2010) A method and server for predicting damaging missense mutations. Nat Methods. 7(4), 248-249.
- 27.Schubach M, Maass T, Nazaretyan L, Röner S, Kircher M. (2024) CADD v1.7: using protein language models, regulatory CNNs and other nucleotide-level scores to improve genome-wide variant predictions. , Nucleic Acids Research 52, 1143-1154.
- 28.Devarajan S, Moon I, Ho M F, Larson N B, Neavin D R et al. (2019) and disposition: the biological fate of chemicals. Pharmacogenomic Next-Generation DNA Sequencing: Lessons from the Identification and Functional Characterization of Variants of Unknown Significance in CYP2C9 and CYP2C19. Drug metabolism 47(4), 425-435.
- 29.Liu D, Ho M-F, Schaid D J, Scherer S E, Kalari K et al. (2017) Breast cancer chemoprevention pharmacogenomics: Deep sequencing and functional genomics of the ZNF423 and CTSO genes. npj Breast Cancer. 3(1), 30.
- 30.Hendrikx T, Porsch F, Kiss M G, Rajcic D, Papac-Miličević N et al. (2022) Soluble TREM2 levels reflect the recruitment and expansion of TREM2+ macrophages that localize to fibrotic are as and limit NASH. , Journal of Hepatology 77(5), 1373-1385.
- 31.Filipello F, Morini R, Corradini I, Zerbi V, Canzi A et al. (2018) . The Microglial Innate Immune Receptor TREM2 Is Required for Synapse Elimination and Normal Brain Connectivity. Immunity 48(5), 979-991.
- 32.Schoch K M, Ezerskiy L A, Morhaus M M, Bannon R N, Sauerbeck A D et al. (2021) Acute Trem2 reduction triggers increased microglial phagocytosis, slowing amyloid deposition in mice. Proceedings of the National Academy of Sciences 118-27.
- 33.B van Lengerich, Zhan L, Xia D, Chan D, Joy D et al. (2023) A TREM2-activating antibody with a blood–brain barrier transport vehicle enhances microglial metabolism in Alzheimer’s disease models. Nature Neuroscience. 26(3), 416-429.
- 34.Liu C, Yu J. (2019) Genome-Wide Association Studies for Cerebrospinal Fluid Soluble TREM2 in Alzheimer's Disease. Frontiers in aging neuroscience. 11, 297.
- 35.Hou X H, Bi Y L, Tan M S, Xu W, Li J Q et al. (2019) Genome-wide association study identifies Alzheimer's risk variant in MS4A6A influencing cerebrospinal fluids TREM2 levels. Neurobiol Aging. 84(241), 213-241.
Cited by (2)
This article has been cited by 2 scholarly works according to:
Citing Articles:
Hami Hemati, Madison B. Blanton, H.E. True, Jude Koura, Rupak Khadka et al. - Brain, behavior, and immunity (2025) Semantic Scholar